ILLUMINA BEADSTUDIO FREE DOWNLOAD
Besides proprietary vendor-provided software BeadStudio, GenomeStudio and open-source software Illuminaio [ 4 ], several pre-processing and quality control QC methods for Illumina bead arrays are available beadarray [ 5 ]; lumi [ 6 ]; limma [ 7 ]. The user may choose to perform all pre-processing steps within our workflow recommended , or to provide already background-subtracted data. To handle chrX, one need to add ,-chrx to the command line parameters and run PennCNV again from there. With the Bookmark Viewer see below , we can examine the details of the CNV calls and export them as text files for further processing. Platform independent web-based Programming language: It cannot detect inserts. 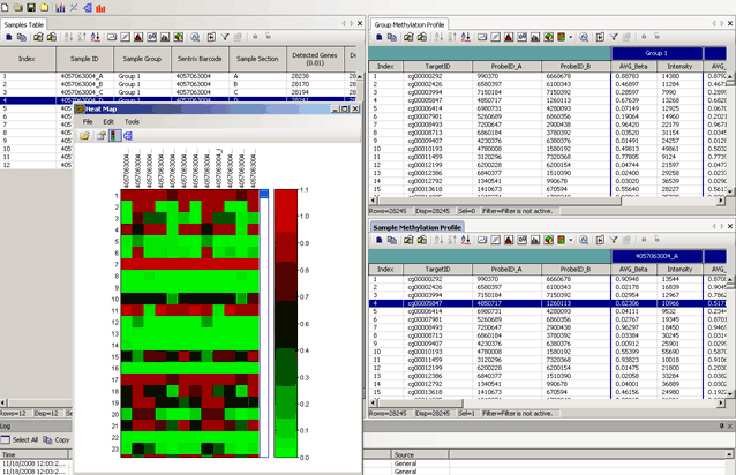
| Uploader: | Vorisar |
| Date Added: | 6 April 2011 |
| File Size: | 30.36 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 17805 |
| Price: | Free* [*Free Regsitration Required] |
Access customized plug-ins and algorithms that can parse and analyze array data exported from GenomeStudio. It enables you to view CpG island methylation status across the genome with the llumina Genome Browser and Illumina Chromosome Browser.
I am familiar with he subprocess module. All files are in binary format. National Center for Biotechnology InformationU.
PennCNV Plug-in - PennCNV
The outputs are directly linked to the existing statistics module of ArrayAnalysis. I've done a few Google searches and haven't come up with anything.
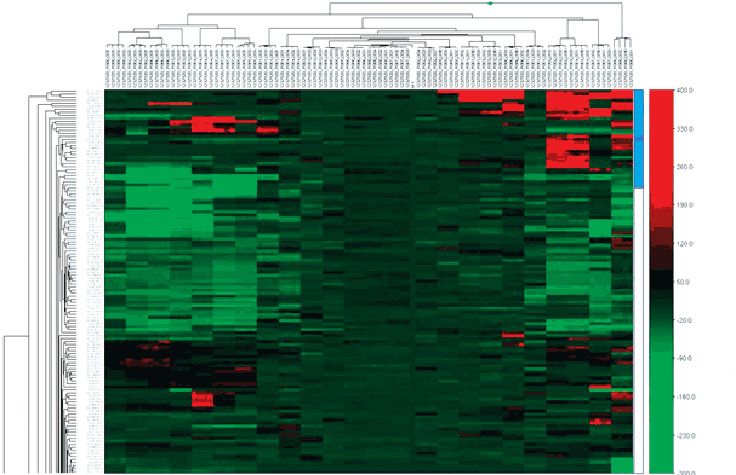
However, the figure below used Human genome build 35, resulting in small discordances. We have recently conducted 1. GT Module Highlights Analyze SNP and CNV data across 5 million markers Estimate Log R ratio and B-allele frequency for copy number analysis Call genotypes, normalize and cluster data, and generate SNP statistics Export genotype data to various third party applications; access multiple CNV algorithms and copy number variation analysis tools Generate bedstudio chromosomal heat map for examining copy number aberrations across the entire genome for multiple samples Analyze data from two different product versions within the same project.
Also, what kind of input does GenomeStudio expect?
Hi all using Illumnia GenomeStudio 2. Funding bodies did not have any role in study design, collection, analysis and interpretation of data, in the writing of the manuscript, and in the decision to submit the manuscript for publication. Data columns are put in matrices in the assayData slot of the resulting object, while annotation fields are put in the featureData slot of the object. Furthermore, open data repositories e. Please review our privacy policy.
Being an open source project, developers within the user community can contribute by adding modules or improving functionality of existing ones, and source code can be downloaded for local deployment.
GenomeStudio Software Downloads
Illuminx are called for each sample dot by their signal intensity norm R and Allele Frequency Norm Theta relative to canonical cluster positions dark shading for a given SNP marker. This encompasses the removal of per-array technical effects, which ensures that the values being further analyzed reflect underlying biology.
Open in a separate window. As in all GenomeStudio modules, the GenomeStudio Framework displays data output in tabular form and enables you to visualize your results quickly and easily using the Illumina Genome Viewer and Illumina Chromosome Browser graphical tools.
Theoretically, if the second application in the pipeline can read from stdinsimply have Python print the required input this goes to stdout and the downstream application can beadstudko this.
Results from the statistics module can then be used for further pathway analysis processing in a downstream module that makes automated calls to PathVisio [ 15 ] or they can be downloaded for processing in other software.
Microarrays Sequencing Popular Genomics Applications. I received genome-wide association GWAS data from a colleague who's supposedly done all the imp One example in BeadStudio is shown below.
GenomeStudio Software Downloads
The Polyploid Genotyping Module supports genotyping data analysis of polyploid organisms such as wheat and potato. Polyploid Genotyping Module Analyze polyploid organism genotyping data View More The Polyploid Genotyping Module supports genotyping data analysis of polyploid organisms such as wheat and potato. This module also enables you to combine methylation data with gene expression profiling experiments within the same GenomeStudio project for correlation between levels of methylated sites beta values and differential gene expression levels p values.
Platform independent web-based Programming language: I am still researching this but I'm hoping someone out there can give me some direction. Kitts and Nevis St. BeadArray expression analysis using bioconductor. bfadstudio
Hi, I am not familiar with beadsrudio, and in that case with command line, but got a script from a c Methylation Module Detect cytosine methylation at single-base resolution Identify methylation signatures across the entire genome View More The GenomeStudio Methylation M module supports the analysis of Infinium and GoldenGate methylation array data.

Comments
Post a Comment